-Search query
-Search result
Showing 1 - 50 of 71 items for (author: de & jong & e)

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-16144: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
Method: single particle / : Hurdiss DL, Drulyte I

PDB-8bon: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
Method: single particle / : Hurdiss DL

EMDB-29714: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Method: single particle / : Ma MW, Hunkeler M, Jin CY, Fischer ES

PDB-8g46: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2
Method: single particle / : Ma MW, Hunkeler M, Jin CY, Fischer ES

EMDB-26547: 
Mediator-PIC Early (Tail A + Upstream DNA & Activator)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26551: 
Mediator-PIC Early (Composite Model)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uik: 
Mediator-PIC Early (Tail A + Upstream DNA & Activator)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uio: 
Mediator-PIC Early (Composite Model)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26542: 
Core Mediator-PICearly (Copy A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26543: 
Mediator-PIC Early (Tail A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26544: 
Mediator-PIC Early (Core B)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26545: 
Mediator-PIC Early (Mediator A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K
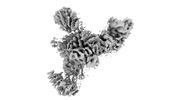
EMDB-26548: 
Mediator-PIC Early (Tail A/B Dimer)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-28023: 
Dimerized Mediator-PIC with early GTFs and TFIIE on a divergent promoter
Method: subtomogram averaging / : Palao III L
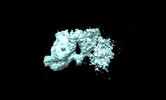
EMDB-28024: 
Dimerized Mediator PIC with early GTFs on a divergent promoter
Method: subtomogram averaging / : Palao III L

PDB-7ui9: 
Core Mediator-PICearly (Copy A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uic: 
Mediator-PIC Early (Tail A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uif: 
Mediator-PIC Early (Core B)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uig: 
Mediator-PIC Early (Mediator A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uil: 
Mediator-PIC Early (Tail A/B Dimer)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-28551: 
RMC-5552 in complex with mTORC1 and FKBP12
Method: single particle / : Tomlinson ACA, Yano JK

PDB-8era: 
RMC-5552 in complex with mTORC1 and FKBP12
Method: single particle / : Tomlinson ACA, Yano JK
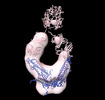
EMDB-26217: 
Negative stain EM map of COVA1-07 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB
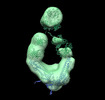
EMDB-26218: 
Negative stain EM map of COVA2-14 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB

EMDB-26219: 
Negative stain EM map of COVA2-18 mAb bound to the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB
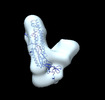
EMDB-26220: 
Negative stain EM map of the S2 domain of SARS-CoV-2 S
Method: single particle / : Han J, Ward AB

EMDB-13139: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with fragment 166 at the transient P-pocket
Method: single particle / : Singh AK, Das K

EMDB-13156: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with fragment F04 at the transient P-pocket
Method: single particle / : Singh AK, Das K

PDB-7ozw: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with fragment 166 at the transient P-pocket
Method: single particle / : Singh AK, Das K

PDB-7p15: 
Cryo-EM structure of HIV-1 reverse transcriptase with a DNA aptamer in complex with fragment F04 at the transient P-pocket
Method: single particle / : Singh AK, Das K

EMDB-11814: 
The cryo-EM structure of the Vag8-C1 inhibitor complex
Method: single particle / : Johnson S, Lea SM, Deme JC, Furlong E, Dhillon A

PDB-7akv: 
The cryo-EM structure of the Vag8-C1 inhibitor complex
Method: single particle / : Johnson S, Lea SM, Deme JC, Furlong E, Dhillon A

EMDB-22989: 
Structure of kinase and Central lobes of yeast CKM
Method: single particle / : Li YC, Chao TC

EMDB-22990: 
Structure of the H-lobe of yeast CKM
Method: single particle / : Li YC, Chao TC

EMDB-22991: 
Structure of the yeast CKM
Method: single particle / : Li YC, Chao TC

PDB-7kpv: 
Structure of kinase and Central lobes of yeast CKM
Method: single particle / : Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL

PDB-7kpw: 
Structure of the H-lobe of yeast CKM
Method: single particle / : Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL

PDB-7kpx: 
Structure of the yeast CKM
Method: single particle / : Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL

EMDB-11657: 
CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc
Method: single particle / : Nakane T, Kotecha A, Sente A, Yamashita K, McMullan G, Masiulis S, Brown PMGE, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, Yu L, Karia D, Pechnikova EV, de Jong E, Keizer J, Bischoff M, McCormack J, Tiemeijer P, Hardwick SW, Chirgadze DY, Murshudov G, Aricescu AR, Scheres SHW

PDB-7a5v: 
CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc
Method: single particle / : Nakane T, Kotecha A, Sente A, Yamashita K, McMullan G, Masiulis S, Brown PMGE, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, Yu L, Karia D, Pechnikova EV, de Jong E, Keizer J, Bischoff M, McCormack J, Tiemeijer P, Hardwick SW, Chirgadze DY, Murshudov G, Aricescu AR, Scheres SHW

EMDB-11638: 
Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A
Method: single particle / : Nakane T, Kotecha A, Sente A, Yamashita K, McMullan G, Masiulis S, Brown PMGE, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, Yu L, Karia D, Pechnikova EV, de Jong E, Keizer J, Bischoff M, McCormack J, Tiemeijer P, Hardwick SW, Chirgadze DY, Murshudov G, Aricescu AR, Scheres SHW

PDB-7a4m: 
Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A
Method: single particle / : Nakane T, Kotecha A, Sente A, Yamashita K, McMullan G, Masiulis S, Brown PMGE, Grigoras IT, Malinauskaite L, Malinauskas T, Miehling J, Yu L, Karia D, Pechnikova EV, de Jong E, Keizer J, Bischoff M, McCormack J, Tiemeijer P, Hardwick SW, Chirgadze DY, Murshudov G, Aricescu AR, Scheres SHW

EMDB-11514: 
Molecular pore spanning the double membranes of murine coronaviral (MHV-A59) replication organelles.
Method: subtomogram averaging / : Barcena M, Wolff G

EMDB-11515: 
Molecular pore spanning the double membranes of a mutant murine coronaviral (MHV-A59 GFP-nsp3) replication organelles.
Method: subtomogram averaging / : Barcena M, Wolff G
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
